Circulating LDL-c levels associated with early onset Alzheimer disease
Higher LDL-c associates with EOAD, independently of APOE E4. Novel rare genetic coding variants were found in APOB, which do not fully explain the association between high LDL-c and EOAD.
Association of Early-Onset Alzheimer Disease With Elevated Low-density Lipoprotein Cholesterol Levels and Rare Genetic Coding Variants of APOBLiterature - Wingo TS, Cutler DJ, Wingo AP et al., - JAMA Neurol. 2019. doi:10.1001/jamaneurol.2019.0648
Introduction and methods
Early onset Alzheimer disease (EOAD) is a form of AD that manifests before the age of 65 years. Heritability of this rare form is 91% to 100% [1]. AD-causing mutations have been identified in the amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2) genes, but it less clear to what extent these genes contribute to EOAD [2]. With likely less than 10% of incident EOAD cases related to these genes, about 90% of EOAD cases remain unexplained [1-3].
Several lines of evidence have suggested a link between increased risk of late-onset AD (LOAD). This study aimed to study the role of circulating cholesterol in relation to EOAD. EOAD is associated with the apolipoprotein E ε4 allele (APOE E4), which is known to raise circulating cholesterol levels, mostly LDL-c [4, 5]. Elevated midlife cholesterol levels have been found in relation to higher risk of AD and cognitive decline, in epidemiological studies, even after adjusting for APOE E4 [6-12]. Cholesterol-lowering therapy has been associated with a lower AD risk, independently of APOE E4 [13].
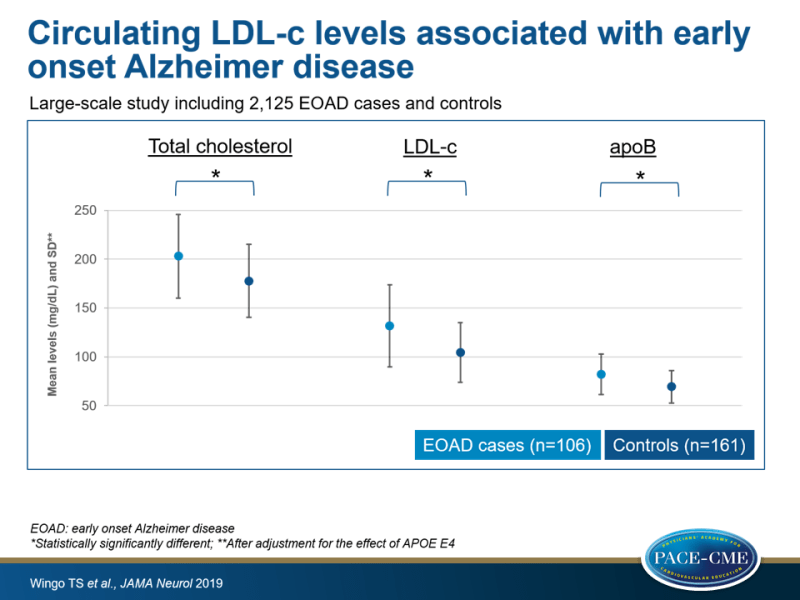
To identify new causes of EOAD, this study examined the contribution of the four mentioned known AD-causing mutations in 2125 EOAD cases and controls. They also investigated whether different fractions of circulating plasma lipoproteins were associated with EOAD after adjusting for APOE E4 in 267 EOAD cases and controls. After finding that EOAD was associated with higher levels of total cholesterol (TC), LDL-c and plasma apolipoprotein B (ApoB), the investigators performed a large-scale targeted sequencing of APOB in the 2125 EOAD cases and controls, as rare variants in this gene are known to strongly affect LDL-c levels [14-16].
EOAD was defined as probable of definite AD based on the National Institute of Neurological and Communicative Diseases and Stroke–Alzheimer’s Disease and Related Disorders Association Work Group criteria [17], with symptoms starting at or before the age of 65 (mean age: 55.6, SD: 4.3). Controls were cognitively normal individuals of 60 years and older (mean age: 72.0, SD: 9.6).
Main results
- Deep sequencing in 2125 samples identified 13 previously described AD-causing mutations, in 23 individuals, among which 2 sites in APP (n=2), 10 sites in PSEN1 (n=18) and 2 sites in PSEN2 (n=3).
- A gene-based burden analysis of rare coding variants in APP, PSEN1 and PSEN2 revealed a significant association only for PSEN1, even though well-described pathogenic mutations were found in all three genes.
- The discovery data set (n=318) showed no enrichment for PSEN1, but larger replication data sets showed a significant association. Further analyses suggested that known AD-causing mutations account for this association. In further analyses to detect new associations, the known AD mutation carriers were excluded.
- 75.1% Of EOAD cases carried the APOE E4 allele, as compared with 30.5% of controls.
- Even after accounting for the effect of APOE E4, EOAD cases had significantly higher mean TC (203.0 [SD: 42.7] vs. 177.7 [SD: 37.2] mg/dL), LDL-c (131.6 [SD: 41.9] vs. 104.4 [30.6] mg/dL) and plasma ApoB (82.2 [SD: 20.8] vs. 69.3 [SD: 16.6] mg/dL) than controls. No significant associations between EOAD and HDL-c or triglyceride levels were seen. It was estimated that LDL-c levels explain 7.6% of the variance in liability to EOAD, independent of APO E4.
- A search for APOB variants associated with coding changes revealed a strong association between APOB and EOAD, independent of APOE E4.
- 57 rare APOB coding variants, 33 of which were found in 31 EOAD cases and 24 in 24 controls (no shared variants) showed that 5% of EOAD patients carried a variant allele, as compared with 1.7% of controls. No evidence was found that particular domains in ApoB were related to LDL-c levels in EOAD cases or controls.
Conclusion
This study found that about 3.4% of all patients with EOAD carry a known AD-causing variant, but only rare coding variants in PSEN1 were associated with EOAD. The strong association between APOE E4 and EOAD was confirmed in this data set. Moreover, a robust association was found between elevated LDL-c levels and EOAD, which was independent of APOE E4. Rare genetic coding variants of APOB were more abundant in EOAD patients than in controls, but this could not fully explain the association between higher LDL-c levels and EOAD, which suggests that additional genes and/or mechanisms are involved.
Editorial comment
In an editorial comment, Makato Ishii [18] note that EOAD patients are often excluded from AD studies, due to the low prevalence of the condition. Because the estimated heritability of EOAD is considerably higher than that of LOAD, and because identified mutations thus far only account for about 11% of EOAD cases, it is likely that there are more AD-susceptibility genes to be discovered. The current study by Wingo et al. confirm that the four tested AD-susceptibility genes account for only a minority of the strong genetic predisposition seen in EOAD.
The study provides the first evidence that rare genetic coding variants of APOB are strongly associated with EOAD, but the data also suggested that there probably are additional contributing factors independent of APOB and APOE. Candidates may include rare variants of other genes involved directly in LDL-c metabolism or factors known to modulate circulating LDL-c levels. Additional studies are needed to further elucidate the association between LDL-c and EOAD. As the authors noted, it is unknown whether protective variants of APOB exist. Identifying such a variant would strengthen the link between APOB and AD pathogenesis. Links between other CV risk factors and EOAD may also be studied and found to be equally enlightening.
Ishii notes that it is a strength of the study that all participants were included in the 29 US Alzheimer's Disease Research Centers (ADRCs), which have standardized research and diagnosis protocols. Nevertheless, some limitations have to be considered, such as that the diagnosis of AD was made by clinical criteria, without use of neuroimaging or cerebrospinal fluid AD biomarkers, allowing for misclassification of EOAD cases that were in fact non-AD dementias. Also, the study population was limited to white people, because of an insufficient number of samples from other ethnic groups. Moreover, factors that influence plasma cholesterol levels, such as dietary intake, drug therapy and hormones, were not taken into account.
Further studies like the one by Wingo et al. should be conducted, not only to identify additional candidate genes involved in AD, but also to discern any differences between EOAD and LOAD that may eventually lead to better management and care of patients with EOAD.
References
1. Wingo TS, Lah JJ, Levey AI, Cutler DJ. Autosomal recessive causes likely in early-onset Alzheimer disease. Arch Neurol. 2012;69(1):59-64. doi:10.1001/archneurol.2011.221
2. Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimers Dement. 2016;12(6): 733-748. doi:10.1016/j.jalz.2016.01.012
3. Campion D, Dumanchin C, Hannequin D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65(3): 664-670. doi:10.1086/302553
4. van Duijn CM, de Knijff P, Cruts M, et al. Apolipoprotein E4 allele in a population-based study of early-onset Alzheimer’s disease. Nat Genet. 1994;7(1):74-78. doi:10.1038/ng0594-74
5. Chasman DI, Paré G, Mora S, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5(11): e1000730. doi:10.1371/journal.pgen.1000730
6. Kivipelto M, Solomon A. Cholesterol as a risk factor for Alzheimer’s disease: epidemiological evidence. Acta Neurol Scand Suppl. 2006;185:50-57. doi:10.1111/j.1600-0404.2006.00685.x
7. Shepardson NE, Shankar GM, Selkoe DJ. Cholesterol level and statin use in Alzheimer disease, I: review of epidemiological and preclinical studies. Arch Neurol. 2011;68(10):1239-1244. doi:10.1001/archneurol.2011.203
8. Kivipelto M, Helkala EL, Laakso MP, et al. Apolipoprotein E 4 allele, elevated midlife total cholesterol level, and high midlife systolic blood pressure are independent risk factors for late-life Alzheimer disease. Ann Intern Med. 2002;137(3): 149-155. doi:10.7326/0003-4819-137-3-200208060-00006
9. Notkola IL, Sulkava R, Pekkanen J, et al. Serum total cholesterol, apolipoprotein E 4 allele, and Alzheimer’s disease. Neuroepidemiology. 1998;17 (1):14-20. doi:10.1159/000026149
10. Kalmijn S, Foley D, White L, et al. Metabolic cardiovascular syndrome and risk of dementia in Japanese-American elderly men: the Honolulu-Asia Aging Study. Arterioscler Thromb Vasc Biol. 2000; 20(10):2255-2260. doi:10.1161/01.ATV.20.10.2255
11. Whitmer RA, Sidney S, Selby J, et al. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology. 2005;64(2):277-281. doi:10.1212/01.WNL.0000149519.47454.F2
12. Jarvik GP, Wijsman EM, Kukull WA et al. Interactions of apolipoprotein E genotype, total cholesterol level, age, and sex in prediction of Alzheimer’s disease: a case-control study. Neurology. 1995;45(6):1092-1096. doi:10.1212/WNL.45.6.1092
13. HaagMD, Hofman A, Koudstaal PJ, et al. Statins are associated with a reduced risk of Alzheimer disease regardless of lipophilicity: the Rotterdam Study. J Neurol Neurosurg Psychiatry. 2009;80(1):13-17. doi:10.1136/jnnp.2008.150433
14. Peloso GM, Auer PL, Bis JC, et al; NHLBI GO Exome Sequencing Project. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014; 94(2):223-232. doi:10.1016/j.ajhg.2014.01.009
15. Lange LA, Willer CJ, Rich SS. Recent developments in genome and exome-wide analyses of plasma lipids. Curr Opin Lipidol. 2015;26(2):96-102. doi:10.1097/MOL.0000000000000159
16. Lange LA, Hu Y, Zhang H, et al; NHLBI Grand Opportunity Exome Sequencing Project. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94(2):233-245.doi:10.1016/j.ajhg.2014.01.010
17. McKhann G, Drachman D, Folstein M et al. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDAWork Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34 (7):939-944. doi:10.1212/WNL.34.7.939
18. Ishii M. Apolipoprotein B as a New Link Between Cholesterol and Alzheimer Disease. JAMA Neurol. 2019. doi:10.1001/jamaneurol.2019.0212
